Functional and Systems Biology
Cells Shown to Multitask in Bacterial Biofilms
A recent study revealed that current concepts of cellular heterogeneity oversimplify bacterial cells’ ability to differentiate into specific cell types and perform tasks.
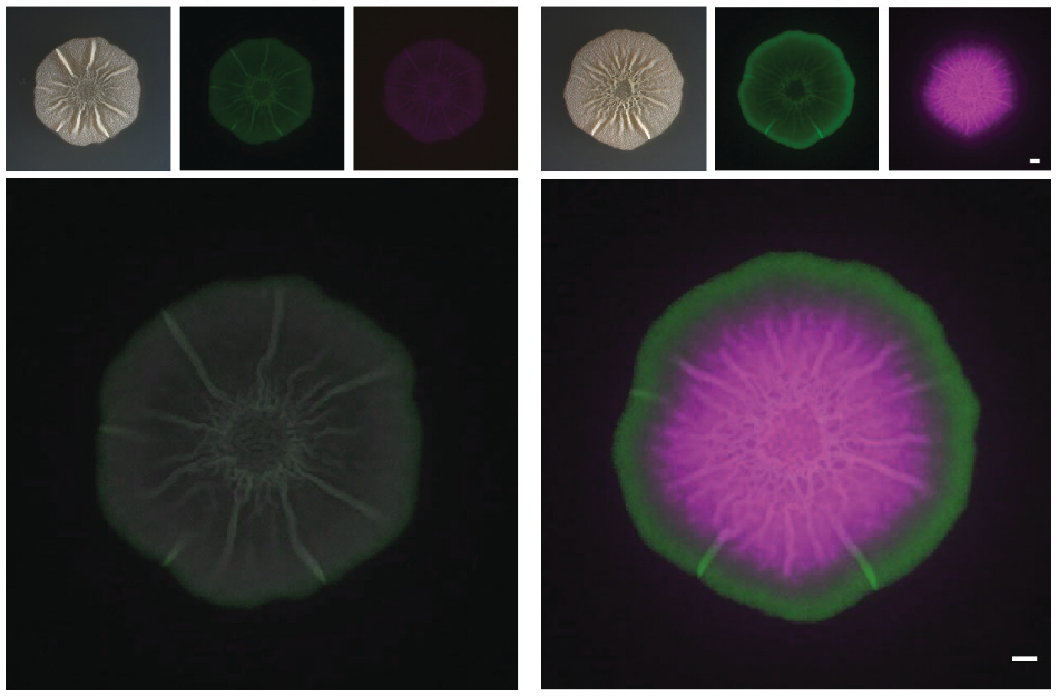
A team of scientists led an experimental study to determine the relationships between subpopulations of cells within the biofilm of a model microbe, revealing new insights regarding their potential. These images show top-down views of colonies of Bacillus subtilis grown on agar plates. The image on the left is a brightfield image while the one on the right shows false-color fluorescence of this colony. These bacterial cells contain two reporter constructs that cause the cells to produce two distinct fluorescence signals (shown as green and pink) when one or both of those genes are expressed. (Image courtesy of Sarah Yannarell | Shank Lab | University of North Carolina, Chapel Hill)
The Science
Bacillus subtilis is a soil-dwelling and biofilm-forming bacterium. Biofilms offer protection to soil microbial communities, while amplifying the ability of cellular communities to carry out important cellular processes that allow them to survive. But knowledge about the phenotypic heterogeneity of the cells and relationships among subpopulations of cells within these biofilms remains largely unknown. A multi-institutional team of scientists led an experimental study to determine the relationships between the subpopulations of cells within these biofilms using the model bacterium B. subtilis. The study revealed that current cellular heterogeneity models oversimplify the ability of bacterial cells to differentiate into specific cell types and perform tasks. Only a few subpopulations of cells appear to have distinct roles, which was surprising given the current understanding of bacterial biofilms.
The Impact
Manipulating the activity of cells within bacterial biofilms requires a greater understanding of cellular differentiation in biofilms. Many microbes express diverse phenotypes to ensure survival in various environments. However, most studies on phenotypic differentiation only examine a few phenotypes at one time, leading to a limited understanding of how cell types overlap and interact across and within the entire biofilm. In this study using the model bacterium B. subtilis, a multi-institutional team investigated the spatial organization and gene expression relationships of reporters for genes important to B. subtilis biofilms. Their findings revealed details about differentiation and phenotypic overlap of cells within mature B. subtilis biofilms. The team mapped spatial gene expression patterns and expanded the number of cell populations described in the literature about B. subtilis. Data from this research provides a framework with which to further study and make predictions about the diverse cellular phenotypes in B. subtilis biofilms.
Summary
A multi-institutional team of scientists generated 182 strains of B. subtilis containing two fluorescent transcriptional reporters to visualize co-localization and quantify co-expression of cell types within biofilms of this species. In total, 14 different genes in pairwise combinations were monitored, and each gene was associated with a cell subpopulation within each biofilm. From there, the team used confocal microscopy at the Environmental Molecular Sciences Laboratory (EMSL), a U.S. Department of Energy (DOE) Office of Science user facility at Pacific Northwest National Laboratory, to determine the spatial organization of gene expression. This revealed that many fluorescent reporters localized to distinct areas of the biofilm, including some that co-localized and others that did not. EMSL’s flow cytometry capability was also used to quantify reporter co-expression, revealing that many cells multi-task, or simultaneously express two reporters. This data challenges current cellular heterogeneity models that assume bacterial cells largely differentiate into specific cell types to perform singular tasks or functions. Instead, only a few subpopulations of cells appear to have distinct roles based on the set of genes examined.
Contacts
Sarah Yannarell, University of North Carolina, Chapel Hill, sarah@wildmicrobes.com
Elizabeth Shank, University of Massachusetts Chan Medical School, Elizabeth.Shank@umassmed.edu
Christopher Anderton, EMSL, Christopher.Anderton@pnnl.gov
Funding
The research was supported by the National Institutes of Health. A portion of the research was performed on a project award from EMSL, a DOE Office of Science user facility. Some of the material is based upon work supported by the DOE Office of Science, Office of Workforce Development for Teachers and Scientists, Office of Science Graduate Student Research program.
Publications
S.M. Yannarell, et al., “Extensive cellular multi-tasking within Bacillus subtilis biofilms.” mSystems 8(4), e00891-22 (2023). [DOI: 10.1128/msystems.00891-22]

